
When the anomaly data is very few, we need to make good use of F1 Score to help us to figure out which model is better.
plot the data
# import modules
%matplotlib inline
import numpy as np
import matplotlib.pyplot as plt
import scipy.io # used to load the mat files
import scipy.optimize
# load the data into numpy
filename = 'ex8/data/ex8data1.mat'
mat = scipy.io.loadmat(filename)
print mat['X'].shape,mat['Xval'].shape,mat['yval'].shape
(307, 2) (307, 2) (307, 1)
X = mat['X']
Xval = mat['Xval']
yval = mat['yval']
def plotData(myX, newFig = True):
if newFig:
plt.figure(figsize=(8,6))
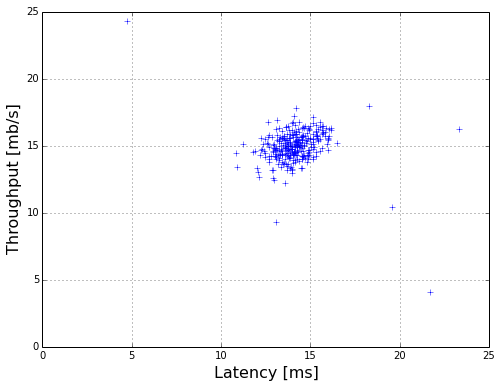
plt.plot(myX[:,0],myX[:,1],'b+')
plt.xlabel('Latency [ms]',fontsize=16)
plt.ylabel('Throughput [mb/s]',fontsize=16)
plt.grid(True)
plt.show()
plotData(X)
cal the mean and deviration
def estimateGaussian(myX):
"""
compute the mean and deviration of the array-like:myX, then return the mean and deviration
"""
mu = np.mean(myX,axis=0)
deviration = myX - mu
dev2 = np.sum(np.power(deviration,2),axis=0)
return mu,dev2 / myX.shape[0]
mu,dev2 = estimateGaussian(X)
print mu.shape, dev2
(2,) [ 1.83263141 1.70974533]
cal the probability
print np.diag(dev2),np.linalg.det(np.diag(dev2))
the output is:
[[ 1.83263141 0. ]
[ 0. 1.70974533]]
3.13333300235
def gaus(myX, mymu, mysig2):
"""
Function to compute the gaussian return values for a feature,
matrix, myX, given the already computed mu vector and sigma matrix.,
If sigma is a vector, it is turned into a diagonal matrix,
Uses a loop over rows; I didn't quite figure out a vectorized implementation.
"""
m = myX.shape[0]
n = myX.shape[1]
if np.ndim(mysig2) == 1:
mysig2 = np.diag(mysig2)
norm = 1.0 / (np.power(2.0 * np.pi, n /2.0) * np.sqrt(np.linalg.det(mysig2)))
myinv = np.linalg.inv(mysig2)
myexp = np.zeros((m,1))
for irow in xrange(m):
xrow= myX[irow]
myexp[irow] = np.exp(-0.5*((xrow-mymu).T).dot(myinv).dot(xrow-mymu))
return norm * myexp
p = gaus(X,mu,dev2)
print p[:10]
the output is:
[[ 0.06470829]
[ 0.05030417]
[ 0.07245035]
[ 0.05031575]
[ 0.06368497]
[ 0.04245832]
[ 0.04790945]
[ 0.03651115]
[ 0.0186658 ]
[ 0.05068826]]
contour the gaussian distribution
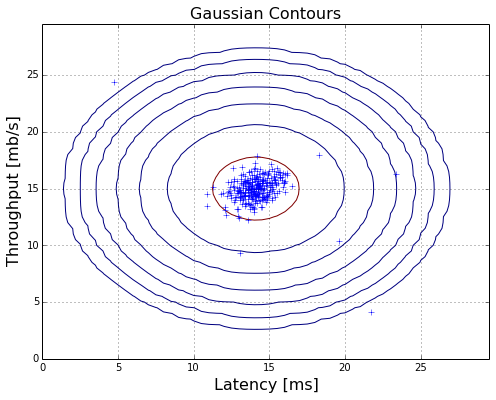
def plotContours(myX, mymu, mysigma2, newFig=False, useMultivariate = True):
delta = 0.5
myx = np.arange(0,30,delta)
myy = np.arange(0,30,delta)
meshx,meshy = np.meshgrid(myx,myy)
coord_list = [ entry.ravel() for entry in (meshx, meshy) ]
points = np.vstack(coord_list)
myz = gaus(points.T,mymu,mysigma2)
#print myz[:,0].shape
myz = myz.reshape((myx.shape[0],myx.shape[0]))
if newFig:
plt.figure(figsize=(8,6))
plt.plot(myX[:,0],myX[:,1],'b+')
plt.xlabel('Latency [ms]',fontsize=16)
plt.ylabel('Throughput [mb/s]',fontsize=16)
plt.grid(True)
cont_levels = [10**exp for exp in range(-20,0,3)]
mycont = plt.contour(meshx,meshy,myz,cont_levels)
plt.title('Gaussian Contours',fontsize=16)
plotContours(X,mu,dev2,True,True)
#plt.show()
F1 score
implementation
def selectThreshold(yval,pval):
"""
yval:label
pval:predict value
function: according to both yval and pval to cal the F1 Score
"""
yval = yval.flatten()
pval = pval.flatten()
bestEpis = 0.0
bestF1 = 0.0
m = len(yval)
step = (np.max(pval) - np.min(pval)) / float(1000)
minv = np.min(pval)
for v in xrange(1000):
episode = minv + v * step
tp = np.sum(np.bitwise_and(pval < episode, yval==1))
fp = np.sum(np.bitwise_and(pval < episode, yval==0))
fn = np.sum(np.bitwise_and(pval > episode, yval==1))
#print tp,fp,fn
if tp + fp ==0:
continue
pre = float(tp)/(tp + fp)
if tp + fn == 0:
continue
rec = float(tp)/(tp + fn)
if (pre + rec)==0:
continue
F1 = 2 * pre * rec/(pre + rec)
if F1 > bestF1:
bestF1 = F1
bestEpis = episode
return bestF1, bestEpis
pval = gaus(Xval,mu,dev2)
bestF1, bestEpis = selectThreshold(yval,pval)
print "best F1 score is:%s,and best episode is:%s)" % ( bestF1, bestEpis )
best F1 score is:0.875,and best episode is:8.99085277927e-05)
visualization
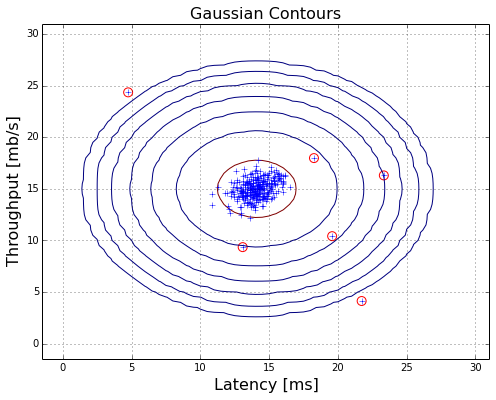
def plotAnomalies(myX,mybestEps, newFig = False):
ps = gaus(myX,*estimateGaussian(myX))
ps = ps.flatten()
anoms = myX[ps < mybestEps]
if newFig:
plt.figure(figsize=(6,4))
plt.scatter(anoms[:,0],anoms[:,1],s=80,facecolors='none',edgecolors='r')
plotContours(X,mu,dev2,True,True)
plotAnomalies(X,bestEpis,False)
#plt.show()
test on 11 dimension samples
filename = 'ex8/data/ex8data2.mat'
mat = scipy.io.loadmat(filename)
X=mat['X']
Xval = mat['Xval']
yval = mat['yval']
print X.shape,Xval.shape,yval.shape
(1000, 11) (100, 11) (100, 1)
mu,sigma2 = estimateGaussian(X)
p = gaus(X,mu,sigma2)
pval = gaus(Xval,mu,sigma2)
bestF1, bestEpis = selectThreshold(yval,pval)
print 'Best epsilon found using cross-validation:%s', bestEpis
print 'Best F1 found on cross validation set: %s', bestF1
print '#Outliers found: %d' % np.sum(p < bestEpis)
Best epsilon found using cross-validation:%s 1.37722889076e-18
Best F1 found on cross validation set: %s 0.615384615385
#Outliers found: 117




近期评论