
Compile libeemd in windows
Part I: Install EEMD packages
- Install Anaconda
- Create a new environment, e.g., EEMD_ENV
- Use terminal in EEMD_ENV
- Install m2w64-gcc, make, m2w64-pkg-config, and m2w64-gsl by using the terminal:
-
conda install make
-
conda install m2w64-pkg-config
-
conda install -c msys2 m2w64-gsl
-
conda install -c msys2 m2w64-gcc
-
- Clone repos:
-
git clone https://bitbucket.org/luukko/libeemd.git
-
git clone https://bitbucket.org/luukko/pyeemd.git
-
- Goto libeemd:
-
cd libeemd
-
- Create a new folder ‘obj’ manually or in DOS:
mkdir obj
-
Edit Makefile in libeemd:
line 58: Delet or comment “ln -sf [email protected] libeemd.so”. This line is only for creating linkage to specific version lib. It does not work in windows.
ln -sf [email protected] libeemd.so
-
Compile libeemd:
make
It will be warning:
1 [main] make 5020 find_fast_cwd: WARNING: Couldn't compute FAST_CWD pointer. Please report this problem to the public mailing list [email protected] make: uname: Command not found cp src/eemd.h eemd.h make: cp: Command not found Makefile:60: recipe for target `eemd.h' failed make: *** [eemd.h] Error 127Igore the information above. That is only because the cmd “cp src/eemd.h” does not work.
We can copy the file “src/eemd.h” manually.OR remove “eemd.h” in line 30:
all: libeemd.so.$(version) libeemd.a
cd ..
- Install pyeemd:
-
cd pyeemd
-
python setup.py install
-
- Copy libeemd files
- Find out which the pyeemd install folder is, e.g., “~/anaconda/envs/EEMD_ENV/lib/python2.7/site-packages/pyeemd-1.4-py2.7.egg/pyeemd”
- Copy “src/eemd.h”, “libeemd.a”, and “libeemd.so.1.4.1” to pyeemd install folder.
- Rename libeemd.so.1.4.1 to libeemd.so
- Finish
Part II: Examples
Taking an example of global temperature time-series
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import statsmodels.api as sm
import pyeemd
url = 'https://data.giss.nasa.gov/gistemp/graphs/graph_data/Global_Mean_Estimates_based_on_Land_and_Ocean_Data/graph.csv';
data = pd.read_csv(url,skiprows=1)
year = data['Year']
tmp = data['No_Smoothing']
tmp = np.array(tmp)
X = sm.add_constant(year)
model = sm.OLS(tmp, X).fit()
predictions = model.predict(X)
imfs = pyeemd.eemd(tmp)
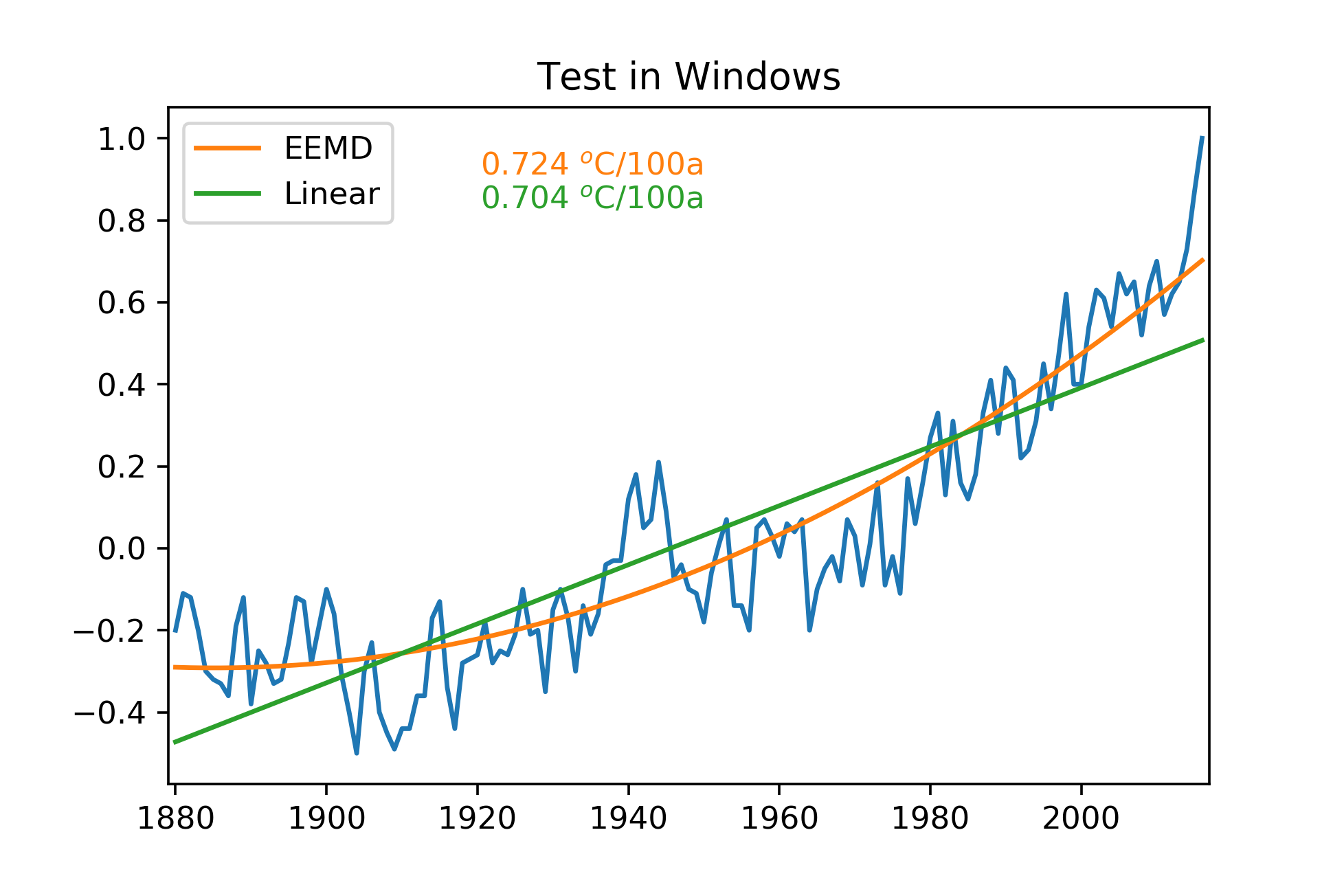
fig = plt.figure()
ax = fig.add_subplot(111)
plt.plot(year, tmp)
eemd_lin = plt.plot(year, imfs[-1,:],label='EEMD')
reg_lin = plt.plot(year, predictions,label='Linear')
plt.legend()
plt.xlim([1879,2017])
trend_eemd = (imfs[-1,-1]-imfs[-1,0])/137.0*100.0
trend_linear = (predictions[134]-predictions[0])/137.0*100.0
plt.text(0.3,0.9, np.str(np.round(trend_eemd,3))+' $^o$C/100a'
, transform=ax.transAxes
, color = eemd_lin[0].get_color())
plt.text(0.3,0.85, np.str(np.round(trend_linear,3))+' $^o$C/100a'
, transform=ax.transAxes
, color = reg_lin[0].get_color())
plt.title('Test in Windows')
plt.savefig('test_win.png',dpi=326)
plt.show()
plt.close()
Part III: Q&A




近期评论